Simulate a deformation field with ANTsR
Brian B. Avants, Jeffrey T. Duda
2020-08-24
deformationSimulation.RmdA binary image defines the “potential” for deformation.
library( ANTsR )
## Loading required package: ANTsRCore##
## Attaching package: 'ANTsRCore'## The following objects are masked from 'package:stats':
##
## sd, var## The following objects are masked from 'package:base':
##
## all, any, apply, max, min, prod, range, sumimg = antsImageRead( getANTsRData( "r16" ) ) ptmat = matrix( c( 95.5, 95.5, 63.5, 63.5 ), ncol=2 ) mymask = img * 0 + 1 aa = makePointsImage( ptmat, mask = mymask, radius = 16 ) aaDist = iMath( aa, "MaurerDistance") + makeImage( mymask, rnorm( sum( mymask ), sd=5 )) plot( aaDist )

## NULLbb = iMath( aa, "MD", 5 ) areg = antsRegistration( aa, aaDist * bb, typeofTransform = "SyNOnly", synMetric = "demons", synIterations = 15, flowSigma = 6, totalSigma = 3, gradStep = 0.2 ) field = antsImageRead( areg$fwdtransforms[ 1 ] ) warpTx = antsrTransformFromDisplacementField( field ) warped = applyAntsrTransform( warpTx, data = img, reference = img) field = antsImageRead( areg$fwdtransforms[ 1 ] ) warpTx = antsrTransformFromDisplacementField( field ) txlist = list( ) ncomp = 6 for ( i in 1:ncomp ) txlist[[ i ]] = warpTx warped = applyAntsrTransform( txlist, data = img, reference = img)
Look.
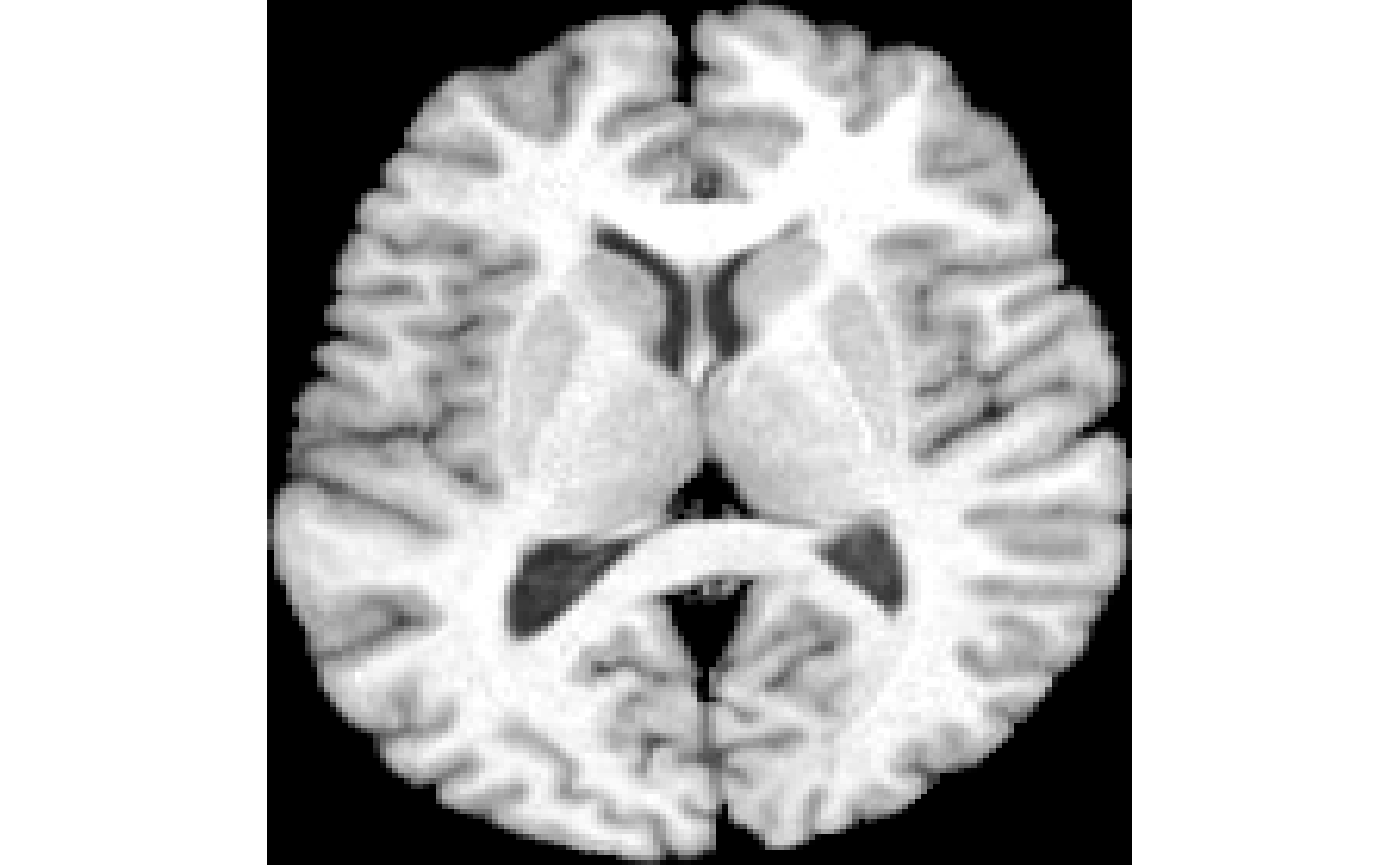
plot( img )
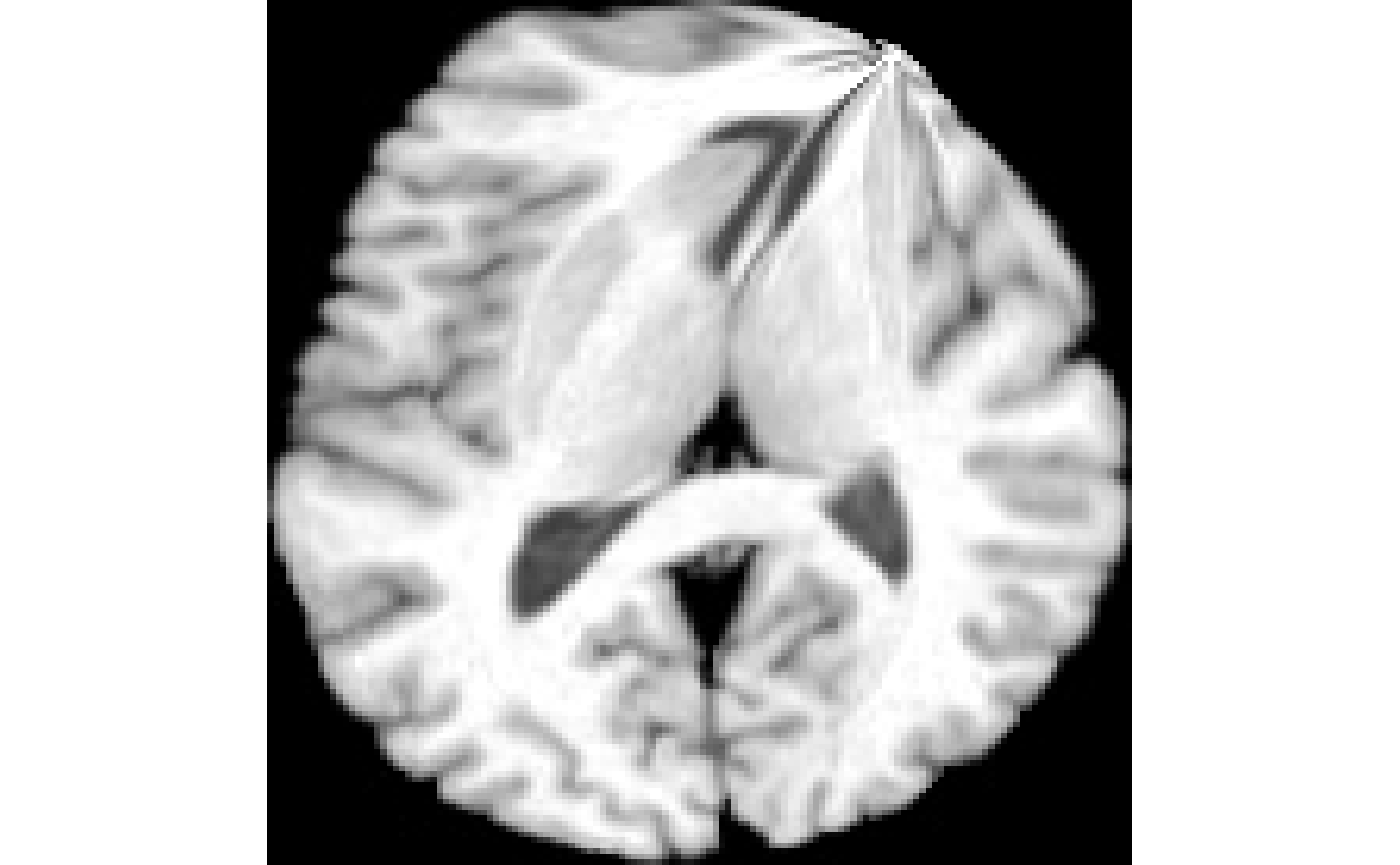
## NULLplot( warped )
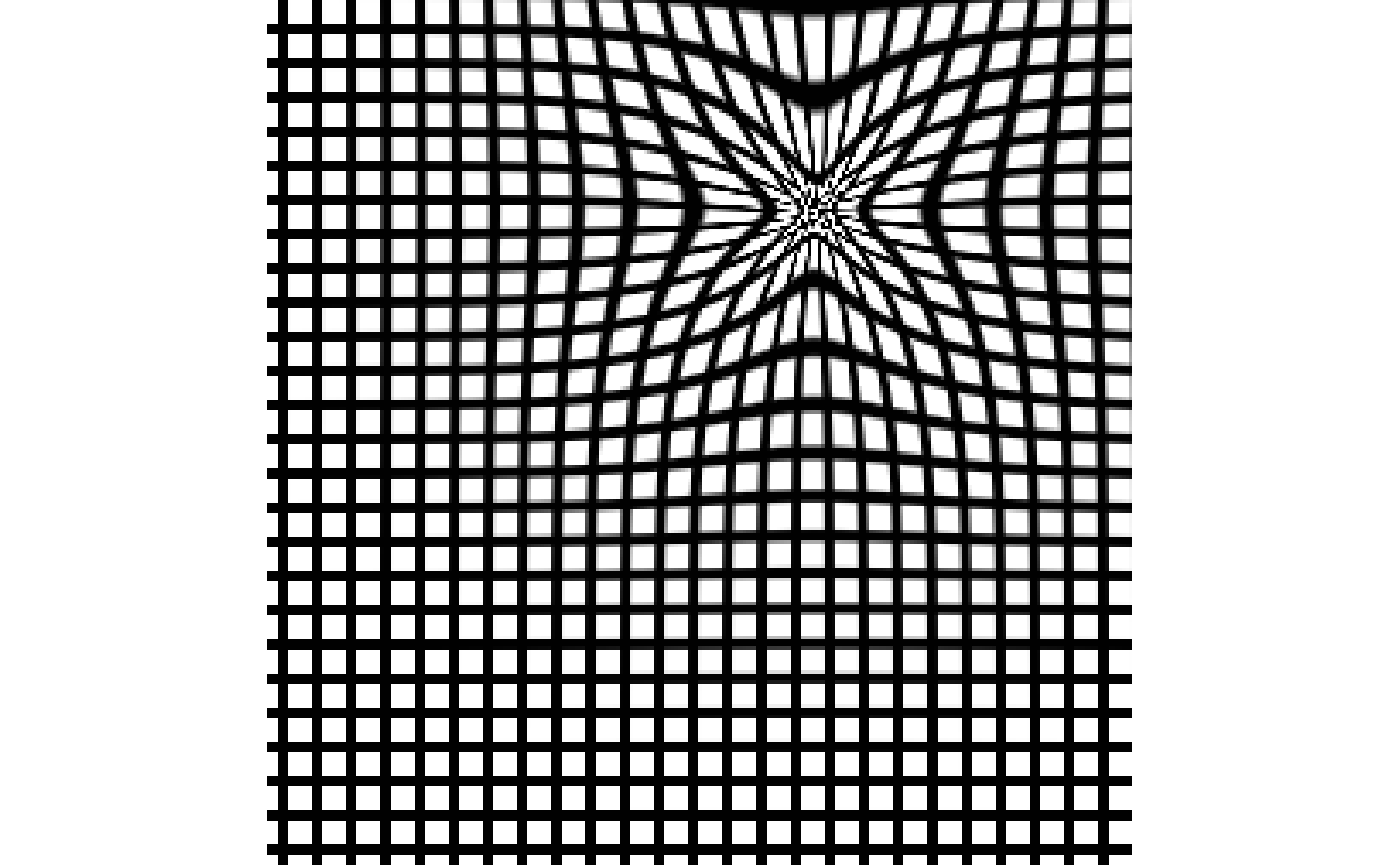
## NULLGrid.
f = areg$fwdtransforms[ 1 ] grid = createWarpedGrid( img, gridStep = 10, gridWidth = 2, fixedReferenceImage = img, transform = rep( f, ncomp ) ) plot( grid )
## NULLCompute a population mean warp.
poplist = list( antsImageRead( getANTsRData( "r16" ) ), antsImageRead( getANTsRData( "r27" ) ), antsImageRead( getANTsRData( "r30" ) ), antsImageRead( getANTsRData( "r62" ) ), antsImageRead( getANTsRData( "r64" ) ), antsImageRead( getANTsRData( "r85" ) ) ) template = antsImageClone( poplist[[ 1 ]] ) if ( !exists( "reglist" ) ) { reglist = list( ) for ( i in 1:length( poplist ) ) { reglist[[ i ]] = antsRegistration( template, poplist[[ i ]], typeofTransform = "SyN" ) } } awrp = antsImageRead( reglist[[ 1 ]]$fwdtransforms[1] ) for ( i in 2:length( reglist ) ) awrp = awrp + antsImageRead( reglist[[ i ]]$fwdtransforms[1] ) awrp = awrp * ( -1.0 * gradstep ) / length( reglist ) warpTx = antsrTransformFromDisplacementField( awrp ) txlist = list( ) ncomp = 10 for ( i in 1:ncomp ) txlist[[ i ]] = warpTx warped = applyAntsrTransform( txlist, data = template, reference = template ) plot( warped )
Curvature flow.
img = antsImageRead( "precuneus.nii.gz" ) msk = thresholdImage( img, 1, Inf ) grad = getNeighborhoodInMask( img, msk, rep(0,img@dimension), get.gradient=T, physical.coordinates = T, boundary.condition = 'mean')$gradients kappa = weingartenImageCurvature( smoothImage(img,0.5) ) * msk # convert grad to channels glist=list() for ( k in 1:3 ) { gradimg1 = msk * 0 gradimg1[ msk == 1 ] = (-1) * grad[k,] * kappa[ msk == 1 ] glist[[ k ]] = gradimg1 } field = mergeChannels( glist ) %>% smoothImage( 1.0 ) jac = createJacobianDeterminantImage( img, field ) warpTx = antsrTransformFromDisplacementField( field * 0.5 / max( abs( field ) ) ) warplist = list( ) for ( i in 1:20 ) warplist[[ i ]] = warpTx warped = applyAntsrTransform( warplist, data=img, reference=img ) # antsImageWrite( warped , '/tmp/temp.nii.gz' )