Creates a multi-row or multi-column plot with image slices and the potential for overlays as well.
multi_overlay(
x,
y = NULL,
z = NULL,
w = 1,
mask = NULL,
col.x = gray(0:64/64),
col.y = hotmetal(),
zlim.x = NULL,
zlim.y = NULL,
ybreaks = NULL,
plane = c("axial", "coronal", "sagittal"),
xlab = "",
ylab = "",
axes = FALSE,
direction = c("horizontal", "vertical"),
par.opts = list(oma = c(0, 0, 0, 0), mar = rep(0, 4), bg = "black"),
text = NULL,
text.x = 0.5,
text.y = 1.4,
text.cex = 2.5,
text.col = "white",
main = NULL,
main.col = text.col,
main.cex = text.cex,
NA.x = TRUE,
NA.y = TRUE,
pdim = NULL,
useRaster = TRUE,
...
)
multi_overlay_center(x, y = NULL, ...)Arguments
| x | List of images of class |
|---|---|
| y | List of images of class |
| z | Slice to display. |
| w | 3D volume to display if |
| mask |
|
| col.x | Color to display x images |
| col.y | Color to display y images |
| zlim.x | Limits for |
| zlim.y | Limits for |
| ybreaks | (numeric) breaks for y to passed to
|
| plane | the plane of acquisition to be displayed |
| xlab | Label for x-axis |
| ylab | Label for y-axis |
| axes | Should axes be displayed |
| direction | Should images be a row or column? Ignored if
|
| par.opts | Options to pass to |
| text | Text to be displayed |
| text.x | Location of text in x-domain |
| text.y | Location of text in y-domain |
| text.cex | Multiplier for text font |
| text.col | Color for |
| main | Title for each plot |
| main.col | Color for |
| main.cex | Multiplier for text font. Will default to |
| NA.x | Should |
| NA.y | Should |
| pdim | Pixel dimensions if passing in arrays. Will be overridden if
|
| useRaster | if |
| ... | Additional arguments to pass to |
Examples
set.seed(5)
dims = rep(10, 4)
arr = array(rnorm(prod(dims)), dim = dims)
arr[,,,c(3, 5)] = rpois(1000*2, lambda = 2)
nim = oro.nifti::nifti(arr)
mask = nim > 2
add_mask = nim[,,,1] > 0
imgs = img_ts_to_list(nim)
masks = img_ts_to_list(mask)
multi_overlay(imgs, masks)
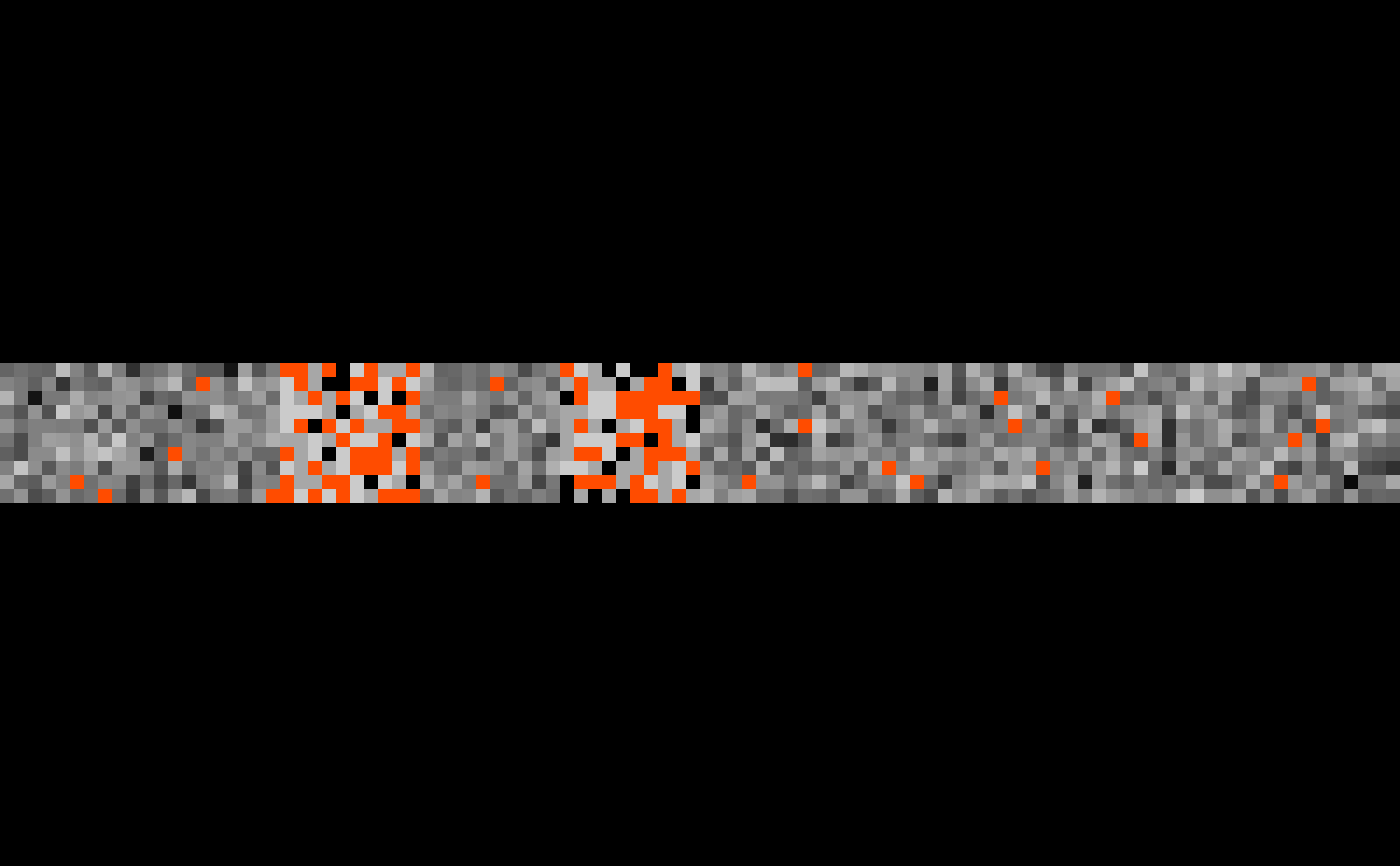 multi_overlay(imgs, masks,
main = "hey", direction = "vertical", plane = "coronal")
multi_overlay(imgs, masks,
main = "hey", direction = "vertical", plane = "coronal")
 multi_overlay(imgs, masks, mask = add_mask,
main = "hey")
multi_overlay(imgs, masks, mask = add_mask,
main = "hey")
 if (FALSE) {
if (requireNamespace("brainR", quietly = TRUE)) {
visits = 1:3
y = paste0("Visit_", visits, ".nii.gz")
y = system.file(y, package = "brainR")
y = lapply(y, readnii)
y = lapply(y, function(r){
pixdim(r) = c(0, rep(1, 3), rep(0, 4))
dropImageDimension(r)
})
x = system.file("MNI152_T1_1mm_brain.nii.gz",
package = "brainR")
x = readnii(x)
mask = x >0
x = lapply(visits, function(tmp){
x
})
alpha = function(col, alpha = 1) {
cols = t(col2rgb(col, alpha = FALSE)/255)
rgb(cols, alpha = alpha)
}
multi_overlay(x, y,
col.y = alpha(hotmetal(), 0.5),
mask = mask,
main = paste0("\n", "Visit ", visits),
text = LETTERS[visits],
text.x = 0.9,
text.y = 0.1,
text.cex = 3)
}
}
if (FALSE) {
if (requireNamespace("brainR", quietly = TRUE)) {
visits = 1:3
y = paste0("Visit_", visits, ".nii.gz")
y = system.file(y, package = "brainR")
y = lapply(y, readnii)
y = lapply(y, function(r){
pixdim(r) = c(0, rep(1, 3), rep(0, 4))
dropImageDimension(r)
})
x = system.file("MNI152_T1_1mm_brain.nii.gz",
package = "brainR")
x = readnii(x)
mask = x >0
x = lapply(visits, function(tmp){
x
})
alpha = function(col, alpha = 1) {
cols = t(col2rgb(col, alpha = FALSE)/255)
rgb(cols, alpha = alpha)
}
multi_overlay(x, y,
col.y = alpha(hotmetal(), 0.5),
mask = mask,
main = paste0("\n", "Visit ", visits),
text = LETTERS[visits],
text.x = 0.9,
text.y = 0.1,
text.cex = 3)
}
}