Adaptive smoothing of ESTATICS parameters and MPM data
smoothESTATICS.RdPerforms adaptive smoothing of parameter maps in the ESTATICS model and if
mpmData is specified these data. Implements both vectorized variants of the
Adaptive Weights Smoothing (AWS, Polzehl and Spokoiny (2006))
and patchwise AWS (PAWS, Polzehl et al (2018)) algorithms with weighting schemes determined by
the estimated parameter maps and their covariances.
smoothESTATICS(mpmESTATICSModel, mpmData = NULL, kstar = 16, alpha = 0.025,
patchsize = 0, wghts = NULL, verbose = TRUE)Arguments
| mpmESTATICSModel | Object of class 'ESTATICSModel' as returned from function |
|---|---|
| mpmData | (optional) Object of class MPMData as created by |
| kstar | Maximum number of steps. |
| alpha | specifies the scale parameter for the adaptation criterion. smaller values are more restrictive. |
| patchsize | Patchsize in PAWS, 0 corresponds to AWS, alternative values are 1 and 2. |
| wghts | (optional) voxel size if measurments are not isotropic. |
| verbose | logical - provide information on progress |
Value
list with components
Estimated parameter maps
map of inverse covariance matrices
convergence indicator map
Sum of weights map from AWS/PAWS
smooting parameters used in AWS/PAWS
smoothed mpmData
image dimension
number of images
vector of T1 filenames
vector of PD filenames
vector of MT filenames
model used (depends on specification of MT files)
filename of brain mask
brain mask
sigma
L
TR values
TE values
Flip angles (FA)
TEScale
dataScale
References
J. Polzehl, V. Spokoiny, Propagation-separation approach for local likelihood estimation, Probab. Theory Related Fields 135 (3), (2006) , pp. 335--362.
J. Polzehl, K. Papafitsorus, K. Tabelow (2018). Patch-wise adaptive weights smoothing. WIAS-Preprint 2520.
J. Polzehl, K. Tabelow (2019). Magnetic Resonance Brain Imaging: Modeling and Data Analysis Using R. Springer, Use R! series. Doi:10.1007/978-3-030-29184-6.
Author
Karsten Tabelow tabelow@wias-berlin.de
J\"org Polzehl polzehl@wias-berlin.de
See also
Examples
# \donttest{
dataDir <- system.file("extdata",package="qMRI")
#
# set file names for T1w, MTw and PDw images
#
t1Names <- paste0("t1w_",1:8,".nii.gz")
mtNames <- paste0("mtw_",1:6,".nii.gz")
pdNames <- paste0("pdw_",1:8,".nii.gz")
t1Files <- file.path(dataDir, t1Names)
mtFiles <- file.path(dataDir, mtNames)
pdFiles <- file.path(dataDir, pdNames)
#
# file names of mask and B1 field map
#
B1File <- file.path(dataDir, "B1map.nii.gz")
maskFile <- file.path(dataDir, "mask.nii.gz")
#
# Acquisition parameters (TE, TR, Flip Angle) for T1w, MTw and PDw images
#
TE <- c(2.3, 4.6, 6.9, 9.2, 11.5, 13.8, 16.1, 18.4,
2.3, 4.6, 6.9, 9.2, 11.5, 13.8,
2.3, 4.6, 6.9, 9.2, 11.5, 13.8, 16.1, 18.4)
TR <- rep(25, 22)
FA <- c(rep(21, 8), rep(6, 6), rep(6, 8))
#
# read MPM example data
#
library(qMRI)
mpm <- readMPMData(t1Files, pdFiles, mtFiles,
maskFile, TR = TR, TE = TE,
FA = FA, verbose = FALSE)
#
# Estimate Parameters in the ESTATICS model
#
modelMPM <- estimateESTATICS(mpm, method = "NLR")
#> Design of the model:
#> [,1] [,2] [,3] [,4]
#> [1,] 1 0 0 0.023
#> [2,] 1 0 0 0.046
#> [3,] 1 0 0 0.069
#> [4,] 1 0 0 0.092
#> [5,] 1 0 0 0.115
#> [6,] 1 0 0 0.138
#> [7,] 1 0 0 0.161
#> [8,] 1 0 0 0.184
#> [9,] 0 1 0 0.023
#> [10,] 0 1 0 0.046
#> [11,] 0 1 0 0.069
#> [12,] 0 1 0 0.092
#> [13,] 0 1 0 0.115
#> [14,] 0 1 0 0.138
#> [15,] 0 0 1 0.023
#> [16,] 0 0 1 0.046
#> [17,] 0 0 1 0.069
#> [18,] 0 0 1 0.092
#> [19,] 0 0 1 0.115
#> [20,] 0 0 1 0.138
#> [21,] 0 0 1 0.161
#> [22,] 0 0 1 0.184
#> done
#> Start estimation in 11200 voxel at 2021-05-17 15:22:50
#>
|
| | 0%
|
|====== | 9%
|
|============ | 18%
|
|=================== | 27%
|
|========================= | 36%
|
|=============================== | 45%
|
|====================================== | 54%
|
|============================================ | 62%
|
|================================================== | 71%
|
|======================================================== | 80%
|
|============================================================== | 89%
|
|===================================================================== | 98%
#> Finished estimation 2021-05-17 15:23:10
#
# smooth maps of ESTATICS Parameters
#
setCores(2)
#> Total CPU cores available: 2 CPU cores in use: 2.
modelMPMsp1 <- smoothESTATICS(modelMPM,
kstar = 16,
alpha = 0.004,
patchsize=1,
verbose = TRUE)
#> using lambda= 125.0501 patchsize= 1
#> Progress:step 1 hakt 1.010096 time 2021-05-17 15:23:10
#> 0.72% mean(bi) 1.11 step 2 hakt 1.022114 time 2021-05-17 15:23:10
#> 1.6% mean(bi) 1.19 step 3 hakt 1.036736 time 2021-05-17 15:23:10
#> 2.8% mean(bi) 1.31 step 4 hakt 1.055091 time 2021-05-17 15:23:10
#> 4.2% mean(bi) 1.46 step 5 hakt 1.079247 time 2021-05-17 15:23:10
#> 5.9% mean(bi) 1.66 step 6 hakt 1.113579 time 2021-05-17 15:23:11
#> 8.2% mean(bi) 1.91 step 7 hakt 1.169869 time 2021-05-17 15:23:11
#> 11% mean(bi) 2.28 step 8 hakt 1.301844 time 2021-05-17 15:23:11
#> 14% mean(bi) 2.96 step 9 hakt 1.432987 time 2021-05-17 15:23:11
#> 19% mean(bi) 3.62 step 10 hakt 1.46909 time 2021-05-17 15:23:11
#> 24% mean(bi) 4.03 step 11 hakt 1.523812 time 2021-05-17 15:23:11
#> 31% mean(bi) 4.62 step 12 hakt 1.627073 time 2021-05-17 15:23:11
#> 39% mean(bi) 5.54 step 13 hakt 1.781336 time 2021-05-17 15:23:12
#> 50% mean(bi) 6.7 step 14 hakt 1.97111 time 2021-05-17 15:23:12
#> 63% mean(bi) 7.99 step 15 hakt 2.236947 time 2021-05-17 15:23:12
#> 79% mean(bi) 9.43 step 16 hakt 2.333957 time 2021-05-17 15:23:12
#> 100% mean(bi) 9.88
#
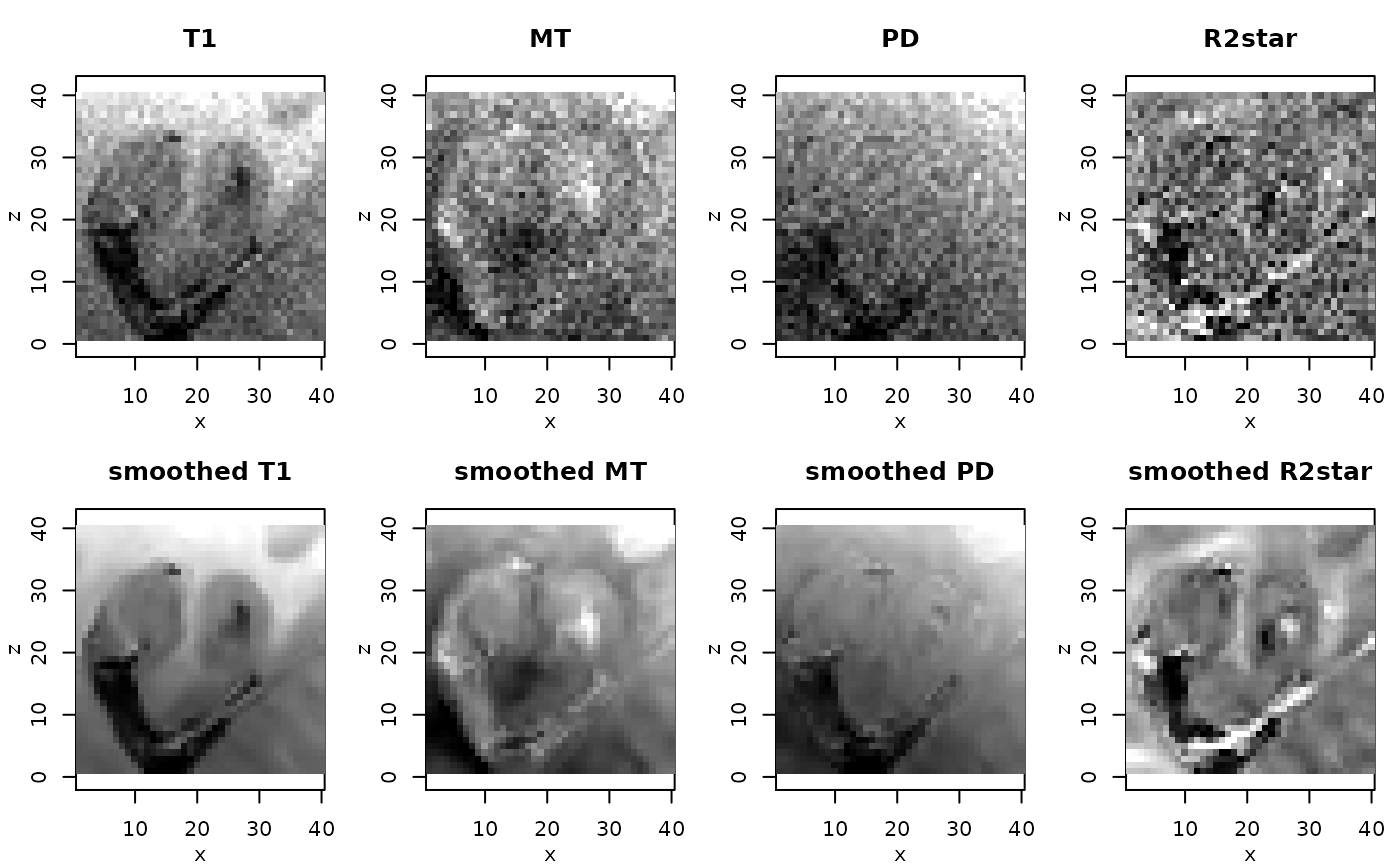
# resulting ESTATICS parameter maps for central coronal slice
#
if(require(adimpro)){
rimage.options(zquantiles=c(.01,.99), ylab="z")
oldpar <- par(mfrow=c(2,4),mar=c(3,3,3,1),mgp=c(2,1,0))
pnames <- c("T1","MT","PD","R2star")
modelCoeff <- extract(modelMPM,"modelCoeff")
for(i in 1:4){
rimage(modelCoeff[i,,11,])
title(pnames[i])
}
modelCoeff <- extract(modelMPMsp1,"modelCoeff")
for(i in 1:4){
rimage(modelCoeff[i,,11,])
title(paste("smoothed",pnames[i]))
}
}
 par(oldpar)
# }
par(oldpar)
# }