This function saves a NIfTI-class object to a single binary file in NIfTI format.
# S4 method for nifti
writeNIfTI(
nim,
filename,
onefile = TRUE,
gzipped = TRUE,
verbose = FALSE,
warn = -1,
compression = 6
)
# S4 method for niftiExtension
writeNIfTI(
nim,
filename,
onefile = TRUE,
gzipped = TRUE,
verbose = FALSE,
warn = -1,
compression = 6
)
# S4 method for anlz
writeNIfTI(
nim,
filename,
onefile = TRUE,
gzipped = TRUE,
verbose = FALSE,
warn = -1,
compression = 6
)
# S4 method for array
writeNIfTI(
nim,
filename,
onefile = TRUE,
gzipped = TRUE,
verbose = FALSE,
warn = -1,
compression = 6
)Arguments
| nim | is an object of class |
|---|---|
| filename | is the path and file name to save the NIfTI file (.nii) without the suffix. |
| onefile | is a logical value that allows the scanning of single-file
(.nii) or dual-file format (.hdr and .img) NIfTI files (default =
|
| gzipped | is a character string that enables exportation of compressed
(.gz) files (default = |
| verbose | is a logical variable (default = |
| warn | is a number to regulate the display of warnings (default = -1).
See |
| compression | The amount of compression to be applied when writing a
file when |
Value
Nothing.
Details
The writeNIfTI function utilizes the internal writeBin and
writeChar command to write information to a binary file.
Current acceptable data types include
- list("UINT8")
DT BINARY (1 bit per voxel)
- list("INT16")
DT SIGNED SHORT (16 bits per voxel)
- list("INT32")
DT SINGED INT (32 bits per voxel)
- list("FLOAT32")
DT FLOAT (32 bits per voxel)
- list("DOUBLE64")
DT DOUBLE (64 bits per voxel)
- list("UINT16")
DT UNSIGNED SHORT (16 bits per voxel)
Methods
- object = "anlz"
Convert ANALYZE object to class
niftiand write the NIfTI volume to disk.- object = "array"
Convert array to class
niftiand write the NIfTI volume to disk.- object = "nifti"
Write NIfTI volume to disk.
References
NIfTI-1
http://nifti.nimh.nih.gov/
See also
Author
Brandon Whitcher bwhitcher@gmail.com,
Volker Schmid
volkerschmid@users.sourceforge.net
Examples
norm <- dnorm(seq(-5, 5, length=32), sd=2)
norm <- (norm-min(norm)) / max(norm-min(norm))
img <- outer(outer(norm, norm), norm)
img <- round(255 * img)
img[17:32,,] <- 255 - img[17:32,,]
img.nifti <- nifti(img) # create NIfTI object
fname = file.path(tempdir(), "test-nifti-image-uint8")
writeNIfTI(img.nifti, fname, verbose=TRUE)
#> writing data at byte = 352
## These files should be viewable in, for example, FSLview
## Make sure you adjust the min/max values for proper visualization
data <- readNIfTI(fname, verbose=TRUE)
#> fname = /tmp/RtmpfdMn90/test-nifti-image-uint8
#> files = /tmp/RtmpfdMn90/test-nifti-image-uint8.nii.gz
#> nii = /tmp/RtmpfdMn90/test-nifti-image-uint8.nii.gz
#> vox_offset = 352
#> seek(fid) = 352
#> # reorient = Method 1
#> ## performPermutation
#> trans =
#> [,1] [,2] [,3]
#> [1,] 1 0 0
#> [2,] 0 1 0
#> [3,] 0 0 1
#> sum(T != 0) = 3
#> det(T) = 1
#> sum(T != 0) == 3 && det(T) != 0 is TRUE
#> ## else {
#> ## real.dimensions = 32 32 32
#> ## dim(out) = 32 32 32
image(img.nifti, oma=rep(2,4), bg="white")
image(data, oma=rep(2,4), bg="white")
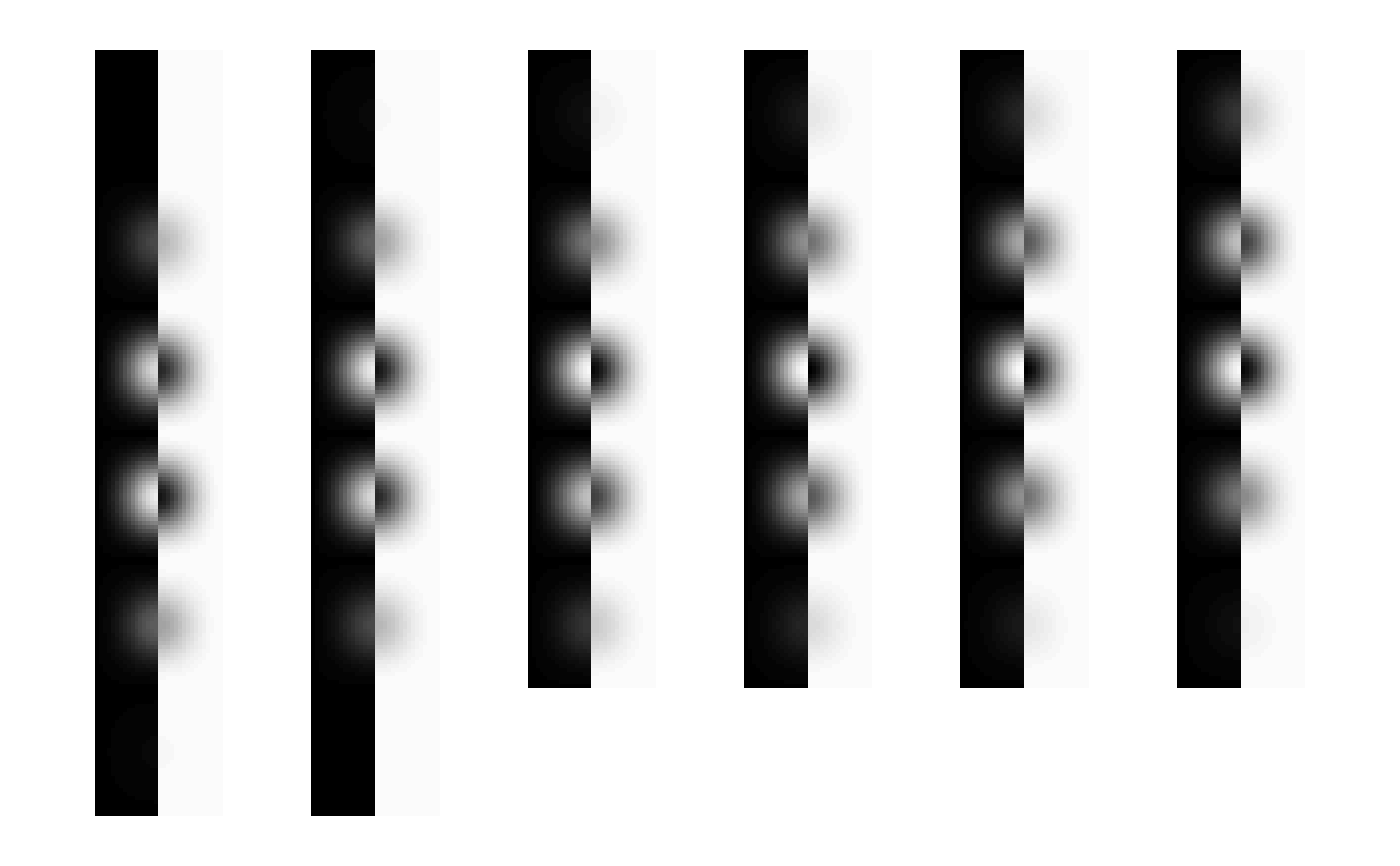 abs.err <- abs(data - img.nifti)
image(as(abs.err, "nifti"), zlim=range(img.nifti), oma=rep(2,4),
bg="white")
abs.err <- abs(data - img.nifti)
image(as(abs.err, "nifti"), zlim=range(img.nifti), oma=rep(2,4),
bg="white")
 if (FALSE) {
## Loop through all possible data types
datatypes <- list(code=c(2, 4, 8, 16, 64),
name=c("uint8", "int16", "int32", "float", "double"))
equal <- vector("list")
for (i in 1:length(datatypes$code)) {
fname <- paste("test-nifti-image-", datatypes$name[i], sep="")
fname = file.path(tempdir(), fname)
rm(img.nifti)
img.nifti <- nifti(img, datatype=datatypes$code[i])
writeNIfTI(img.nifti, fname, verbose=TRUE)
equal[[i]] <- all(readNIfTI(fname) == img)
}
names(equal) <- datatypes$name
unlist(equal)
}
if (FALSE) {
## Loop through all possible data types
datatypes <- list(code=c(2, 4, 8, 16, 64),
name=c("uint8", "int16", "int32", "float", "double"))
equal <- vector("list")
for (i in 1:length(datatypes$code)) {
fname <- paste("test-nifti-image-", datatypes$name[i], sep="")
fname = file.path(tempdir(), fname)
rm(img.nifti)
img.nifti <- nifti(img, datatype=datatypes$code[i])
writeNIfTI(img.nifti, fname, verbose=TRUE)
equal[[i]] <- all(readNIfTI(fname) == img)
}
names(equal) <- datatypes$name
unlist(equal)
}