OASIS Prediction
oasis_predict.RdThis function creates the OASIS probability map from a single MRI study with FLAIR, T1, T2, and PD volumes.
oasis_predict(flair, t1, t2, pd = NULL, brain_mask = NULL, model = NULL,
return_preproc = FALSE, binary = FALSE, threshold = 0.16,
verbose = TRUE, ...)Arguments
| flair | flair volume of class |
|---|---|
| t1 | t1 volume of class |
| t2 | t2 volume of class |
| pd | pd volume of class |
| brain_mask | brain mask of class |
| model | an object of class |
| return_preproc | is a logical value that indicates whether the
preprocessed images should be returned, if |
| binary | logical indicating whether a binary map should be returned by thresholding the probability map |
| threshold | numeric indicating the threshold value for the probability map, with default of 0.16 for the OASIS paper |
| verbose | print diagnostic messages |
| ... | options passed to |
Value
A list of volumes:
the OASIS probability map, the preprocessed volumes (if
return_preproc = TRUE),
the brain mask for the subject, the voxel selection mask, and a thresholded,
binary mask (if binary = TRUE) .
Examples
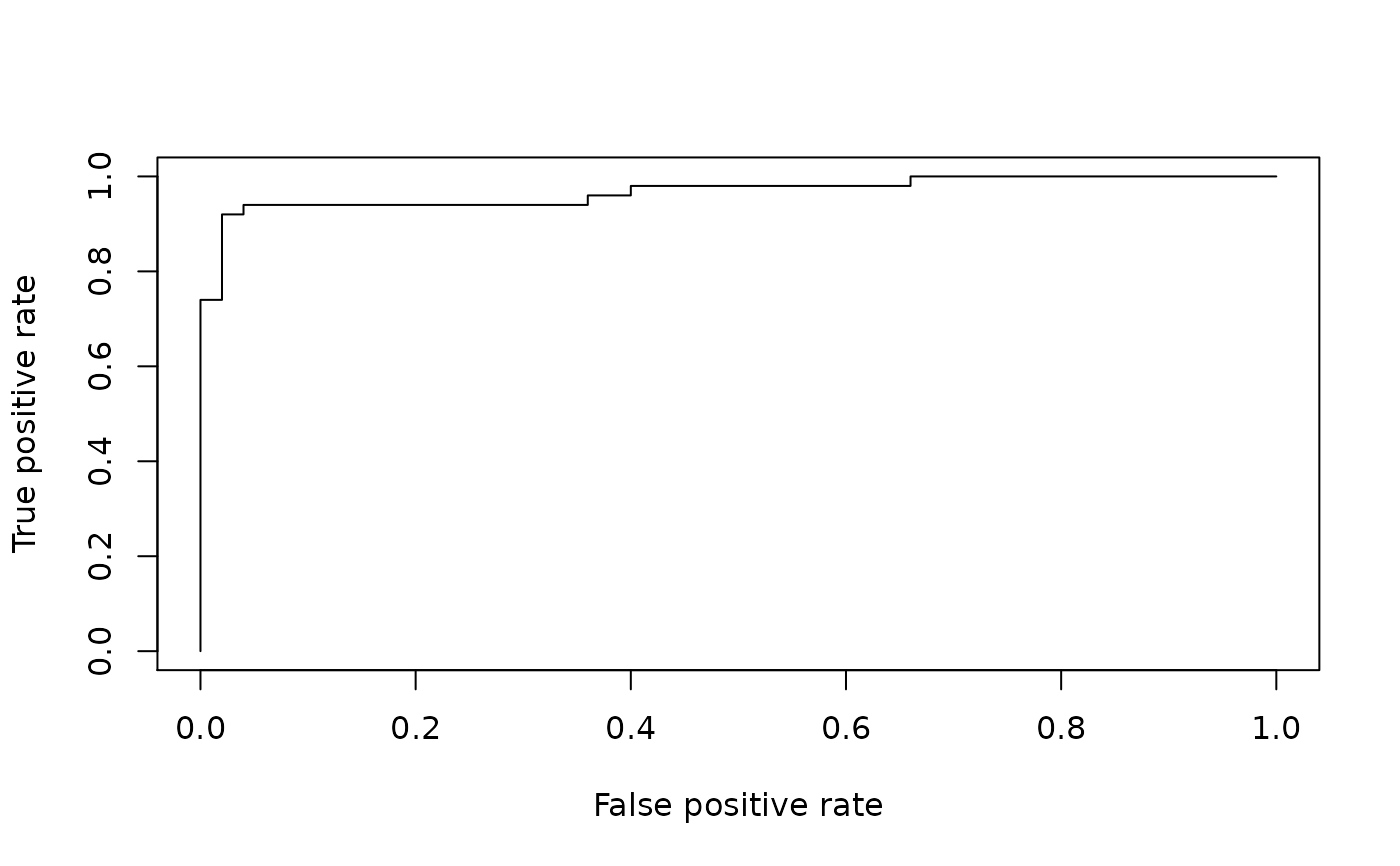
library(ROCR)
p = predict( oasis::oasis_model,
newdata = example_oasis_df,
type = 'response')
nopd_p = predict( oasis::nopd_oasis_model,
newdata = example_oasis_df,
type = 'response')
y = example_oasis_df$GOLD_Lesions
pred = ROCR::prediction(p, y)
perf = ROCR::performance(pred, "tpr", "fpr")
plot(perf)
 library(neurobase)
dl_file = function(url) {
tfile = tempfile(fileext = ".nii.gz")
req <- httr::GET(url,
httr::write_disk(path = tfile))
httr::stop_for_status(req)
tfile
}
in_ci <- function() {
nzchar(Sys.getenv("CI"))
}
on_cran = function() {
identical(Sys.getenv("NOT_CRAN"), "false")
}
if (in_ci() || on_cran()) {
if (fslr::have.fsl() && require(httr)) {
mods = c("FLAIR", "T1W", "T2W", "consensus_gt", "brainmask")
base_url = file.path(
"https://raw.githubusercontent.com/muschellij2/open_ms_data",
"master/cross_sectional/coregistered/patient01/")
files = paste0(base_url, mods, ".nii.gz")
files = sapply(files, dl_file)
names(files) = mods
flair <- readnii(files["FLAIR"])
t1 <- readnii(files["T1W"])
t2 <- readnii(files["T2W"])
brain_mask <- readnii(files["brainmask"])
gold_standard = readnii(files["consensus_gt"])
oasis_preprocessed_data <- oasis_predict(flair, t1, t2,
brain_mask = brain_mask, preproc = TRUE)
}
}
library(neurobase)
dl_file = function(url) {
tfile = tempfile(fileext = ".nii.gz")
req <- httr::GET(url,
httr::write_disk(path = tfile))
httr::stop_for_status(req)
tfile
}
in_ci <- function() {
nzchar(Sys.getenv("CI"))
}
on_cran = function() {
identical(Sys.getenv("NOT_CRAN"), "false")
}
if (in_ci() || on_cran()) {
if (fslr::have.fsl() && require(httr)) {
mods = c("FLAIR", "T1W", "T2W", "consensus_gt", "brainmask")
base_url = file.path(
"https://raw.githubusercontent.com/muschellij2/open_ms_data",
"master/cross_sectional/coregistered/patient01/")
files = paste0(base_url, mods, ".nii.gz")
files = sapply(files, dl_file)
names(files) = mods
flair <- readnii(files["FLAIR"])
t1 <- readnii(files["T1W"])
t2 <- readnii(files["T2W"])
brain_mask <- readnii(files["brainmask"])
gold_standard = readnii(files["consensus_gt"])
oasis_preprocessed_data <- oasis_predict(flair, t1, t2,
brain_mask = brain_mask, preproc = TRUE)
}
}